Signaling, regulation and metabolic interactions
Team MetaboActions / Michael Hodges
Improvement of crop potential requires the identification of specific parts of metabolism, which can be manipulated to achieve better outputs. When nutrient supply is such that existing yield potential is reached, the only way to increase production is to improve the efficiency by which nutrients are taken up and used. Metabolism may be manipulated either to achieve more carbon assimilation (per unit of nitrogen) at a lower energetic cost or to increase the biosynthesis of energy compounds (for example NAD) and/or the capacity of nutrient use. An understanding of the basic processes of metabolic pathways and how they relate to plant biomass and respond to environmental constraints is of importance to guide plant breeding and to drive genetic engineering.
The MetaboActions team has 6 current major research interests:
- Regulation of photorespiratory cycle enzymes.
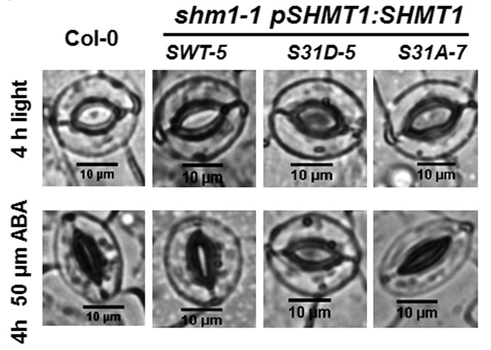
- Role of photorespiration in stomatal movements.
- Energy metabolism: NAD biosynthesis and recycling.
- Beneficial microbes to increase plant growth under climate change conditions.
- Acclimation of primary metabolism to climate change conditions.
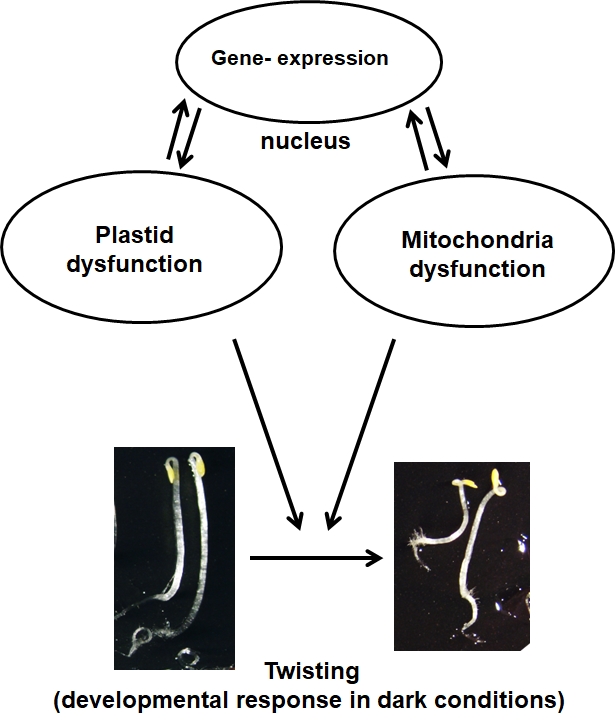
- Impact of organelle dysfunction on seedling development: skotomorphogenesis & retrograde signalling

Regulation of photorespiratory enzymes (Mathieu Jossier, Nathalie Glab)
Photorespiration begins when Rubisco assimilates O2 instead of CO2 to produce “toxic” 2-phosphoglycolate (2PG) and “useful” 3-phosphoglycerate (3PGA). The photorespiratory cycle metabolizes 2PG to form 3PGA but in doing so it removes assimilated carbon and nitrogen to produce CO2 and ammonium while consuming energy. Deemed a wasteful process, photorespiration has been a target to improve photosynthesis and plant yield. To date, the core photorespiratory genes and enzymes have been identified and characterized but little is known about how the photorespiratory cycle and its interactions with neighbouring metabolic pathways are regulated.
The recent development of proteomics has led to a wealth of data concerning post-translational modifications (PTM). This has suggested that all photorespiratory enzymes can be phosphorylated and redox regulated. Over the last few years, we have been trying to decipher the function and role of identified phosphorylation sites associated with Arabidopsis thaliana phosphoglycolate phosphatase (PGLP1), glycolate oxidase (GOX1 & GOX2), serine hydroxymethyltransferase (SHMT1) and hydroxypyruvate reductase (HPR1). More recently, we have turned our attention to the potential redox regulation of PGLP1 that has been reported to be sensitive to H202, and associated with methionine oxidation and nitrosyation.
We are following two major strategies: The in vitro analysis of PTM-site mutated recombinant proteins and the functional complementation of photorespiratory mutant lines with synthetic genes to produce either phosphorylation-mimic, or non-phosphorylatable or redox-insensitive photorespiratory enzymes.
Role of photorespiration in stomatal movements (Mathieu Jossier)
Photorespiration is a metabolic pathway inherent to photosynthesis, strongly influenced by climate change and which in turn can greatly affect photosynthesis. Indeed, the energy cost of photorespiration and its competition with photosynthetic CO2 assimilation has a negative impact on plant growth and yield. This phenomenon will be intensified by climate change, due to higher temperatures and more frequent periods of drought. In this context, the control of stomatal opening is crucial and improving the response of stomata to environmental changes could greatly improve photosynthesis and water use efficiency, thus supporting plant productivity. Photorespiration has recently been implicated in modifying CO2-dependent and light-dependent stomatal movements and conductance but the underlying mechanism(s) is/are still unknown. The objectives of this project are to decipher the role of photorespiration in stomatal movements in Arabidopsis thaliana and to determine if this involves the photorespiratory activity of stomatal guard cells and/or leaf mesophyll cells. The guard cell signaling pathway(s) and metabolic mechanisms involved in photorespiration-influenced stomatal movements are also examined. This project is based on the study of photorespiratory mutants of Arabidopsis thaliana mainly using gas exchange measurements, microscopy and metabolic profiling by GC-MS.
Energy metabolism: NAD biosynthesis and recycling (Bertrand Gakière)
We are also interested in understanding the interactions between NAD and plant primary metabolisms. Despite the important role of NAD in plant metabolism and stress signalling, little is known about NAD synthesis, except for the identification of at least two possible pathways. To test the importance of NAD in plant metabolism, we have identified and characterized several key Arabidopsis thaliana NAD biosynthesis/recycling genes and proteins including L-aspartate oxidase, quinolinate phosphoribosyltransferase and nicotinate/ nicotinamide mononucleotide adenyl transferase (N(a)MNAT.
A better understanding of the biochemical properties of recombinant L-aspartate oxidase and N(a)MNAT has provided new knowledge of how NAD biosynthesis might be regulated by these key enzymes. In parallel, the importance of these genes on NAD homeostasis, pathogen defence, abiotic stress resistance, growth and development are investigated in T-DNA insertion mutants and overexpressing plants. In this way a correlation was observed between plant NAD content and aspartate oxidase expression and activity.

The industrial value of this work has been protected (De Bont & Gakière, EU & US patent application) and an IDEX Prematuration grant from the University Paris-Saclay (EnergyCrop) has allowed us to begin testing the overexpression of Arabidopsis L-aspartate oxidase in tomato, colza and rice.
Beneficial microbes to increase plant growth under climate change conditions (Axel de Zélicourt)
The constant release of CO2 in the atmosphere is a major component of global climate change. In addition, this increase profoundly modifies the physiology of plants, in particular by acting on major metabolic pathways such as photosynthesis, photorespiration as well as the assimilation and use of nitrogen. Beneficial bacteria interact with plants, improving their growth and tolerance to environmental stresses. Enterobacter sp. SA187 is a beneficial bacterium that increases plant tolerance to abiotic stresses (Andrès-Barrao et al., 2017; de Zélicourt et al., 2018; Shekawat et al., 2021). Recently, we have shown that this beneficial bacterium also improves the growth of Arabidopsis thaliana when cultivated under elevated CO2 conditions (eCO2, 1000 ppm). Therefore, we seek to determine and characterize which pathways are modulated by Enterobacter sp. SA187 and lead to this beneficial effect on plant growth under eCO2 conditions. Using the plant model Arabidopsis thaliana, and by combining physiological measurements with omics analyses (transcriptomics and metabolomics), we aim to answer the following questions:
- What is the influence of SA187 inoculation on plant development and physiology under eCO2 conditions?
- What are the plant and bacterial signaling pathways and metabolic functions (e.g. hormone content, photosynthetic activity, nitrogen use,…) involved in this beneficial interaction?
![SA187 root colonization promotes Arabidopsis growth in e[CO2]. (A) Mock and SA187 inoculated plants grown for 17 days under e[CO2] conditions. (B) Mock and SA187 inoculated plants fresh weight under ambient and e[CO2] conditions. Box plots represents 5 biological replicates (n>70), asterisks indicate a statistical significance based on t-test (*= p<0.05; ***=p<0.0001). (C) Colonization of Arabidopsis root using GFP-labelled SA187 strain.](/_richText-file/ametys-internal%253Asites/ips2/ametys-internal%253Acontents/signalisation-regulation-et-interactions-metaboliques-article-2/_attribute/content/_data/figure%20SA197bis_203x238.jpg)
In addition, we are currently screening a bacterial collection to identify new beneficial bacteria that bring about increased plant growth under eCO2conditions and/or improved tolerance to heat stress. The most promising bacteria will be characterized and tested directly on crops (such as wheat or rapeseed) in order to provide new tools to increase plant yield in the near future.
Acclimation of primary metabolism to climate change conditions (Michael Hodges)
Photosynthesis plays a central role in capturing atmospheric CO2 and therefore it is a key factor to be considered in mitigating the rise in global CO2 levels. The predicted rise in atmospheric CO2 of up to 750-1300 ppm before the end of the century is expected to be accompanied by a rise in temperature. An increase in CO2 should stimulate photosynthetic CO2-fixation by 40% and thus favourably impact biomass production. However, this stimulation is much lower than expected (typically in the range of 0-20%) in many crops including cereals. In addition, elevated CO2 has a negative effect on plant mineral status and plant protein content. The lower-than-expected stimulation of CO2 assimilation and plant growth by elevated CO2 is due to photosynthetic and mineral nutrition acclimations that are still poorly understood. The signalling events and pathways associated to these acclimations to elevated CO2 have yet to be identified and little is known about the combined effect of elevated CO2 and high temperature on CO2-capture and nutrient status. Metaboactions is part of a consortium that hopes to obtain funding (PEPR FairCarboN) to decipher the complexity of plant acclimation to climate change conditions with an aim of improving CO2 capture and storage.
Metaboaction is a partner in an INRAe DIGIT-BIO 2021 project called PEERSIM coordinated by IPS2 GNET/OGE teams and aimed at re-evaluating the prediction of a multi-stress scenario (eCO2 and elevated temperature) based on single stress and double stress multi-omics studies of Arabidopsis. While, the metabolism-metabolome facility (PMM) is a partner of an EIG Concert Japan project (IRUEC) which aims to improve resource use efficiency in cereals, under climate change. In this project wheat and rice cultivars are challenged by high temperature and CO2 levels under different N fertilization conditions.
Impact of organelle dysfunction on seedling development: skotomorphogenesis & retrograde signalling (Livia Merendino)
Plants are sessile organisms and they are forced to cope with changing environmental conditions. Plants are a very good model system to analyse the mechanisms for adapting to the environment and the role played by mitochondria and plastids, the bioenergetics organelles in the cell, in these processes.
We have recently discovered that in response to mitochondrial respiratory limitation the morphogenetic pattern of Arabidopsis plants during early development in the dark is highly modified as to improve resistance to mechanical stress. We are currently investigating how this response could be used by the plant during underground seed germination to facilitate emersion from compact soil. The knowledge coming from these experiments could inspire new strategies to optimize the under-ground development program of cultivated plants and, as direct consequence, to enhance the survival rate of emerging plants from the soil.
Previous research highlights include:
- Identification of C reallocation when the photorespiratory cycle is blocked leading to reduced photosynthetic activity and the biosynthesis of less Rubisco protein to maintain C/N balance; Dellero et al (2015) Plant J & (2016) J Exp Bot. This involved the development of 13C tracing methods coupled to NMR analysis.
- Evidence showing the crucial role of SnRK1 activation/phosphorylation by SnAK kinases in plant development; Guerinier et al (2013), Glab et al (2017) Plant J.
- NAD energy metabolism acts as an integral regulator of multiple defence layers; Petriacq et al (2016) Plant Physiol. Increased NAD levels improve plant biomass and seed yield (De Bont & Gakière Patent).
- Serine 82 phosphorylation is involved in the reaction mechanisms of Arabidopsis thaliana 2,3-bisphosphoglycerate-independent phosphoglycerate mutase 2 (Duminil et al (2021) Plant J).
Major approaches used:
- Reverse genetics: Production and selection of genetically-modified plants (insertion mutants, artificial microRNA, overexpressors, complemented lines with mutated genes).
- Recombinant proteins & enzymology: Production and purification of tagged-recombinant proteins (wild-type or modified by site-directed mutagenesis) and the analysis of their kinetic properties.
- Phenotyping & plant physiology: Analyses including metabolites (mainly primary & redox metabolisms) by GC-MS, LC-MS/MS, HPLC, stable-isotope labeling (13C,15N) coupled to NMR, H+-NMR, gas exchange measurements (CO2 and H2O), enzymatic activities, gene expression analyses (qPCR, RNAseq), and microscopy.
- Translational biology from plant models to crop plants.
The Metabolism-Metabolome facility (Bertrand Gakière, Françoise Gilard)
The PMM is devoted to providing adapted analytical services. A major goal is to offer expertise to analyse plant metabolites by developing isotopic and metabolomics protocols to allow metabolic phenotyping of plant lines. In this way, PMM helps to characterize the consequences of abiotic/biotic stresses and specific mutations, and the determination of carbon, nitrogen and water use efficiencies using the adequate technologies. These include: Semi-targeted and targeted metabolite profiling by GC-MS and NMR; isotope analyses on total organic matter (EA-IRMS), gases (GC-IRMS) and extracts (LC-IRMS); analyses of cofactors (by LC-MS), amino acids (by HPLC) and untargeted metabolomics (combining LC-MS/MS and GC-MS). PMM has contributed to research topics including: water stress and C/N metabolisms in Medicago, cytochrome c oxidase Arabidopsis mutants, CO2 effects on C metabolism in wheat flag leaves, sugar accumulation in melon, natural 13C distribution in oil palm, metabolite profiling of maize inbred lines inoculated with N-fixing bacteria, MAP kinase defense responses and salicylic acid (SA) and C-reallocation in photorespiratoty mutants and enzyme reaction mechanism.
PMM is currently involved in several National and International projects. Ongoing projects include:
- CYTOPHENO (ANR project, F Budard, IJPB INRA Versailles).
- AMAIZING & Project Nitrogen Use Efficiency (“Investissement d'avenir”, B.Hirel, IJPB INRA Versailles).
- PISTILL (SPS LABEX project, A. Boualem, IPS2).
- Orchidomics (EU project, MA Sellosse, MNHN Paris).
- Water stress tolerance (FSOV project, JC Deswartes, Arvalis).
In parallel, PMM is developing mycotoxin detection protocols within the frame of the FUSAKILL project (M. Dufresne, IPS2).
The recent acquisition of a LC-MS/MS will allow PMM to detect molecules that are difficult to measure by GC-MS and LC-MS. It will also open up the possibility to undertake fluxomics analyses after isotopic-labeling. Non-targeted metabolomics is being developed including secondary metabolites that interest the perfume industry (PISTILL & FRAGRANCE, A. Boualem, IPS2), and the rubber industry (CIRAD Montpellier). The platform is also committed to making all data accessible via a public repository (Metabolights) and it is developing new tools to enhance the presentation of results and analytic procedures.
