Stress signaling
Team STRESS / Jean Colcombet and Hannes Kollist
As sessile organisms, plants must adapt to the local perturbations of their environment, i.e. to biotic and abiotic stress. Understanding the ways plants can perceive these stresses and cope with them constitutes a major area of research, notably to engineer more resilient crops for the future.
Kinase signaling is a very efficient and common mean to transmit cellular information. Plant genomes usually encode for several thousands of kinases, which underlines the importance of this type of signaling in this organism. Among the variety of kinase groups, the STRESS team is more specifically interested in elucidating the roles of two central families : the Mitogen-Activated Proteine Kinases (MAPKs or MPKs) and the Calcium-Dependent Protein Kinases (CDPKs or CPKs).
- MAPK modules are composed of a MAP3K, a MAP2K and a MAPK that activate each other by phosphorylation in a serial manner. In plants MAPK modules are encoded by large multigenic families. Most of the current knowledge focuses on MPK3, MPK4 and MPK6 which are the downstream MAPK of several different modules involved in responses to biotic and abiotic stresses as well as in developmental processes. In contrast very little is known about the other members of the MAPK family.
- Plant CDPKs are able to translate nuclear or cytoplasmic fluctuations of calcium concentration into phosphorylation signals. Among the 34 Arabidopsis CDPKs, CPK5 and CPK6 are known to be two key regulators of various stress responses. For instance, in immunity, both kinases act redundantly to regulate oxidative burst, phytoalexin and hormone synthesis, and transcriptional reprogramming. However the underlying molecular mechanisms remain poorly known.
The six research topics of the laboratory
What are the roles of CDPKs in plant adaptation to abiotic stress ?
Marie Boudsocq
The osmotic stress caused by drought, soil salinity or cold is a major environmental constraint affecting plant yield. This stress induces a cell dehydration that comes with an ionic desequilibrium and an accumulation of ROS. To maintain its integrity, plant triggers signaling pathways regulating gene expression, metabolism and antioxydant systems. Calcium contributes to these responses as a second messenger and is decoded by several sensors including CDPKs. We showed that the double mutant cpk5cpk6 is hypersensitive to salt stress. Currently we are developping « omic » approaches to identify the underpinning molecular mechanisms.
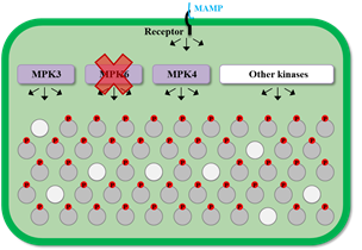
What are the roles of MAPK modules in the regulation of plant immunity ?
Julien Lang, Jean Colcombet
The MPK3/6 module is known to be activated in a rapid and transient fashion during PAMP-Triggered Immunity (PTI) and in a delayed and more sustainable fashion during Effector-Triggered Immunity (ETI). Currently we are investigating how these different kinetics of activation impact plant defense responses, especially at the level of transcriptional reprogramming.
Moreover we recently identified a new MAPK module that is activated in a sustainable fashion during PTI. The objective is now to characterize the role of this new module in the implementation of immune responses, and to analyze its link with the traditional MPK3/6 module.
What are the substrates of MAPKs and CDPKs in stress context ?
Marie Boudsocq, Julien Lang, Jean Bigeard
Identifying subtrates of kinases is essential to unravel the molecular mechanisms that mediate their biological functions. For this we follow two strategies. First we are looking for kinase interactors either through identification of in planta complexes by mass spectrometry (TAP-MS), or through y2H screening. Second we perform some phosphoproteomic analyses, comparing wild-type controls with lines inducibly overexpressing constitutively active forms of the kinases or on the contrary with lines showing loss of functions for specific kinases. Currently we are focusing on three candidates : a chromatin remodeling factor known to be involved in defense responses, as well as an inhibitor of phosphatase and a transcription factor which have been both identified as putative common targets of both MAPKs and CDPKs, and which thereby offer an original opportunity to study the connexions between the two signaling pathways.
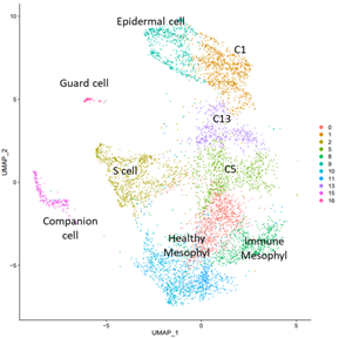
Single cell analysis of plant immune responses
Julien Lang
Plant defense responses involve several biological processes that allow plants to fight against pathogenic attacks. How these different processes are orchestrated within organs and might depend on specific cell types is poorly known. Using scRNAseq technology on Arabidopsis leaves inoculated with the bacterial pathogen Pseudomonas syringae, we are characterizing the immune responses dependent on different cell types (mesophyl cells, epiderm cells, guard cells, and vascular cells). We are also inferring trajectories showing how distinct cells populations might converge towards similar defense responses, or alternatively diverge between different kinds of responses. In addition we are analyzing how a plant mutant defective in MAPK signaling behaves in response to pathogens at the single cell level. Overall our data provide an innovative framework to describe and explore plant cell responses to biotic stress.
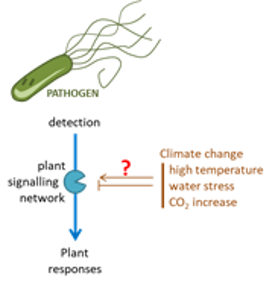
Impact of climate change on biotic stress perception by plants
Jean Colcombet, Erwan Le Deunff
Climate change is likely to affect food security and threaten social organizations worldwide. On top of more numerous heat waves and dryness periods, temperature increase will also favor the poleward migration of tropical pathogens, further jeopardyzing crop yield. To date the number of studies adressing the concomitant effects on plant signaling of heat and biotic stress is limited. Our team recently found that some signaling pathways that are essential for defense responses are shut down by high temperature. We are now trying to get a broader insight on this phenomenon to elucidate its molecular basis.
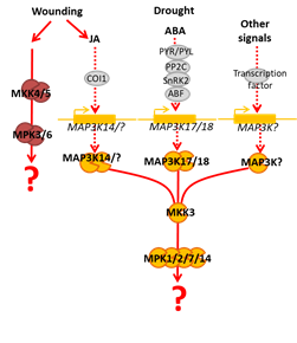
Role of the MKK3 module in integrating various environmental constraints
Jean Colcombet
In 2015 our team reported the finding of a new, complete and atypical MAPK module composed of MAP3K17/18-MKK3-MPK1/2/7/14. This module is activated by ABA and allows plants to adapt to drought. Unlike other MAPK modules which are generally activated within minutes, MPK1/2/7/14 are activated by ABA after several hours. This activation further depends on the de novo synthesis of the MAP3Ks. To our knowledge this was the first description of such an atypical MAPK module in plants as well as in animals. Later we revealed that wounding could also activate MPK1/2/7/14 and that this activation also required MKK3. But in this context, the upstream MAP3K appears to be mainly MAP3K14. Based on these findings we assume that the MKK3-MPK1/2/7/14 module might be involved in response to various environmental cues. Our objective is to identify these cues and to understand how they might contribute to the plant adaptation by mediating specific or general stress responses.