Few examples of applications
Metabolomics analysis with technic of gas chromatography coupled with mass spectrometry (GC-MS)
Collaboration with B. Hirel – IJPB. Identification of a number of maize metabolites representative of plant-bacterial interaction and specific for the strains, which have a functional nitrogenase activity. (Liziane Cristina Brusamarello-Santos, Françoise Gilard, Lenaïg Brulé, Isabelle Quilleré, Benjamin Gourion, Pascal Ratet, Emanuel Maltempi de Souza, Peter J. Lea, Bertrand Hirel : Metabolic profiling of two maize (Zea mays L.) inbred lines inoculated with the nitrogen fixing plant-interacting bacteria Herbaspirillum seropedicae and Azospirillum brasilense. PLoS ONE 03/2017 ; 12(3)., DOI:10.1371/journal.pone.0174576).
Isotopics analysis with technic of elementar analysis coupled with isotopic ratio mass spectrometry (EA-IRMS)
Collaboration with Arvalis for the project FSOV 2012 K : Identification of drought tolerance traits and development of tools to support their evaluation (Jean-Charles DESWARTE, Katia BEAUCHENE, Eric OBER, Thierry MOITTIE, Camille BEDARD,Laure DUCHALAIS, Jérémy DERORY, Céline ZIMMERLI, Valérie LAURENT, Clément DEBITON, Bertrand GAKIERE, Frédéric BARET ; Colloque FSOV 03/2017.
|
|
|
|
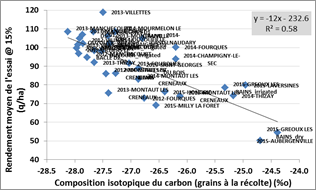
Relationship between isotopic composition of grain carbon at harvest (average value measured on 2 control varieties) and average yields measured in the multilocal test network |
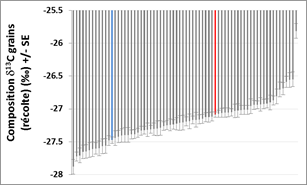
Histogram illustrating the genetic variability (adjusted average) of d13C in the multilocal test network. Error bars indicate the standard error. The colored bars mark the two witnesses Apache (blue) and Bermude (red). |
Fluxomics analysis with technic of nuclear magnetic resonance (NMR)
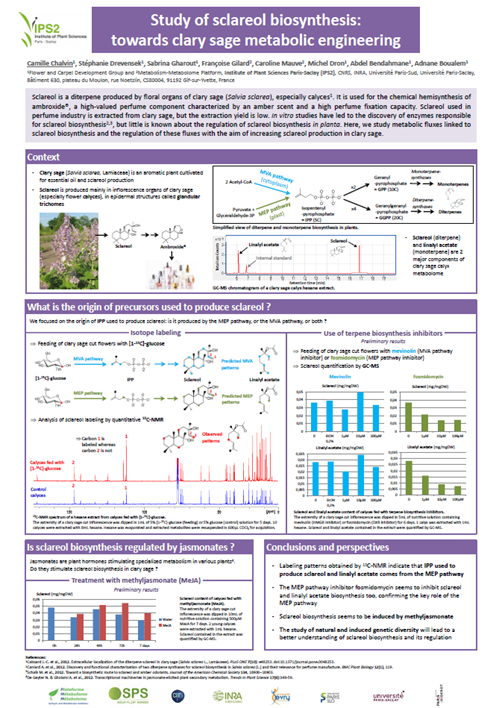
Project SPS2020 PISTILL and pHD Camille Chalvin (collaboration with M. Dron and A. Boulaem –IPS2) : The objective is to dissect the synthesis pathway of a terpenoid obtained from a lamiaceae and which has a strong interest for aromatic industry. Isotopic molecules feeding experiments, analysis by mass spectrometry and fluxomics are used to identify the various intermediate molecules and the metabolic fluxes of the pathway.
Co-factors analysis with technic of liquid chromatography ultra high-performance coupled with mass spectrometry (LC-MS)
NAD Metabolism. Florence Guérard, Pierre Pétriacq, Bertrand Gakière, Guillaume Tcherkez : Liquid chromatography/time-of-flight mass spectrometry for the analysis of plant samples: A method for simultaneous screening of common cofactors or nucleotides and application to an engineered plant line. Plant Physiology and Biochemistry, volume 49, Issue 10, October 2011, p1117-1125.
|
|
|
|
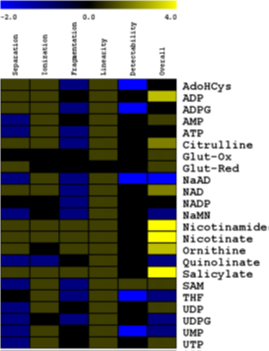
Summary of the LCeTOF MS performance for the analysis of selected metabolites. |
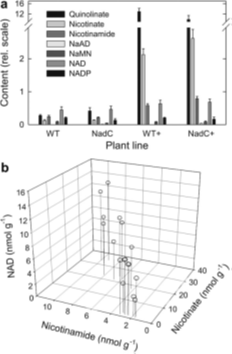
Leaf content in metabolites involved in NAD biosynthesis in wild-type and nadC lines. |
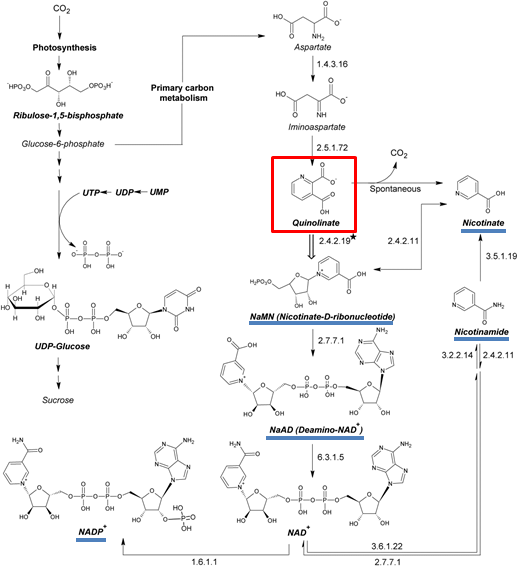
The metabolic pathway investigated here, that includes nicotinamine dinucleotide (NAD) biosynthesis. The engineered Arabidopsis line examined in the present study (nadc) expresses the gene coding for bacterial quinolinate phosphoribosyl transferase (QPRT, EC 2.4.2.19), indicated with a star. Other enzymes are indicated by their EC number.