Bioinformatics
IPS2 informatics core provides resources and skills to support IPS2 teams in the field of Bioinformatics.
- For small scale analysis we have access to local power data center of 276 processors, 780 Go memory and 3 GPU cards.
- For large scale analysis we have access to a Data Center (GENOTOUL) of about 5000 cores (INTEL-2014, AMD-2012), 34 Tera Byte memory (3TB on a SMP machine) and data storage.The data center permits access to more than 200 softwares, web servers and virtual machines hosting infrastructure.
Examples of tools developed by the platform with the support of the IPS2 informatics core.
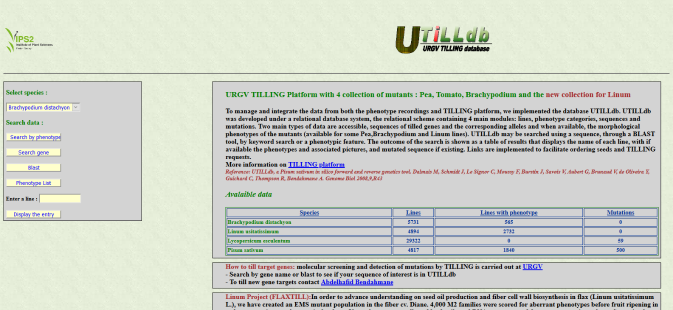
UTILLdb
In order to manage and integrate the expanding data from both the phenotype recordings and TILLING target genes, we implemented the database UTILLdb. UTILLdb was developed according to a relational database system, interconnecting four main modules: lines, phenotype categories, sequences and mutations. Two main types of data are accessible, the morphological phenotypes of mutants and the sequences of tilled genes and corresponding alleles, when available. UTILLdb may be searched using a sequence, through a BLAST tool or for a phenotypic feature using a keyword search. The outcome of the search is shown in a table that displays the phenotype of each line, with pictures and mutated sequence if it exists. Since the phenotypic characterization of the TILLING mutants will become more detailed, UTILLdb was designed so that the passport data of the mutant lines can be extended or modified as needed. UTILLdb manage two types of data: publicly accessible data and confidential data. Accessible data could be searched through a web interface with a link to facilitate seed ordering and serves as an entry point for users wishing to have their favorite gene tilled.
http://tools.ips2.u-psud.fr/UTILLdb
Reference
Dalmais M, Schmidt J, Le Signor C, Moussy F, Burstin J, Savois V, Aubert G, Brunaud V, de Oliveira Y, Guichard C, Thompson R, Bendahmane A. UTILLdb, a Pisum sativum in silico forward and reverse genetics tool. Genome Biol. 2008;9(2):R43
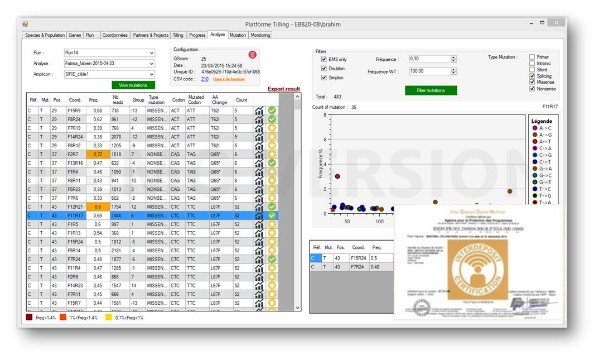
SENTINEL
SENTINEL is a user-friendly software allowing the platform users to analyse NGS sequencing data as well as to archive the results. It combines a bioinformatics analysis pipeline with archive databases.
A new version of SENTINEL, SENTINDEL, enables identification of INDELS.
Bendahmane, A., Marcel F., Dalmais M., Beaumont G., Mania B. SENTINEL, SOFTWARE dedicated to TILLING by NGS Analysis. Certifier par l’Agence pour la Protection des programmes. Inter Deposit Digital Number.FR001.240004.000.R.P.2016.000.10000
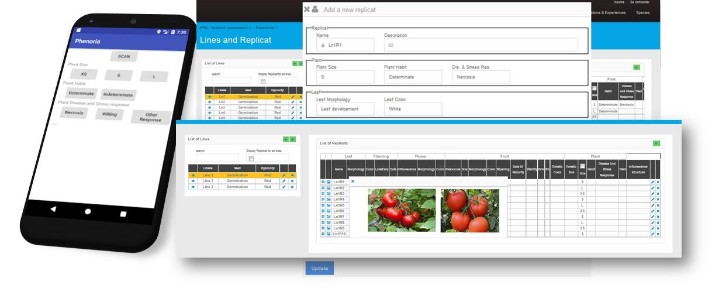
PHENORIA
PHENORIA is an Android application for data collection via mobile computers. It was developed on the platform to facilitate the collection and the organisation of phenotyping data on mutant collections, regardless of the production location. Users can create an experiment and select the best phenotyping ontology. The file can be shared with other users.
Reference
Mania B. Gomez V. Boualem A. Bendahmane, A. PHENORIA, an Android based app for managing Plant Phenotypic Data.
