Genome wide analysis of NL genes in common bean genome
Team GDYNPATH / Valerie Geffroy
Analyses of small RNA, DNA methylation, and genome sequence of common bean, using new sequencing technologies with a specific focus on NL disease resistance genes and satellite DNAs
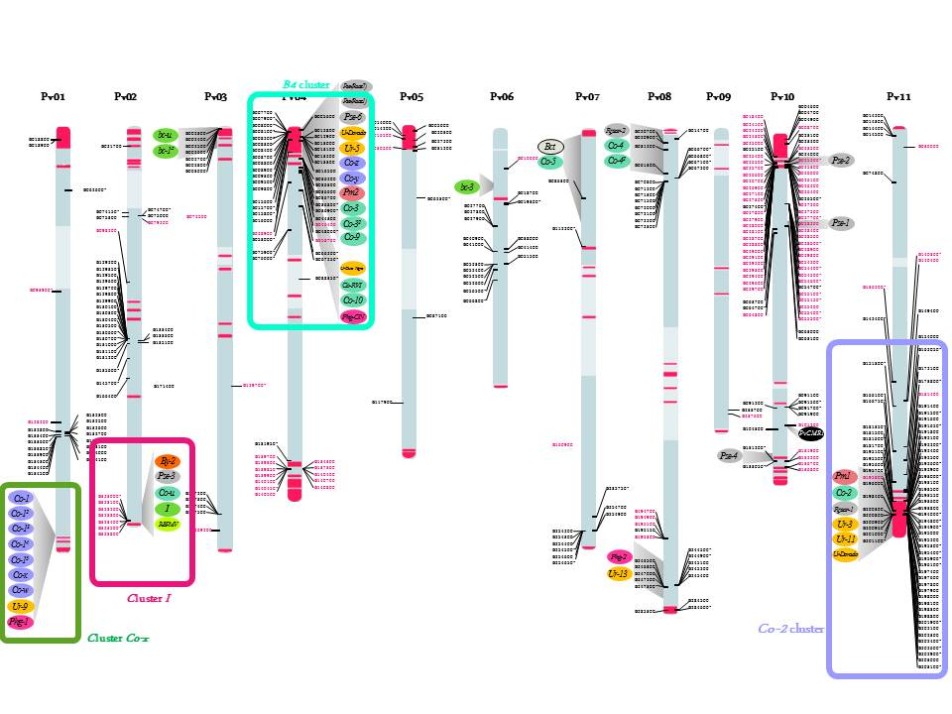
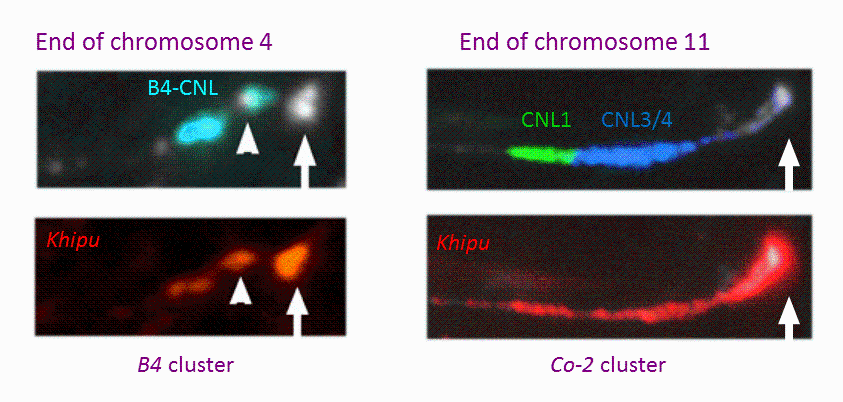
Previously considered as an orphan crop, important genomic resources are now available in common bean thanks to the advent of next-generation sequencing (NGS) technologies. In particular, the genome sequence of common bean, G19833 an Andean landrace, has recently been assembled by a consortium led by S.A. Jackson (UGA; USA) (Schmutz et al., 2014). Our group was involved in this project via the annotation of NL sequences. Access to common bean genome sequence provides unparalleled insight into the organization and evolution of this large multigene family in P. vulgaris. We identified ~400 NLs and, as observed in other plant species, most of them are organized in complex cluster of genes with the particularity to be often located in subtelomeric regions close to terminal knobs containing the satellite DNA khipu (Richard et al. 2013, Ribeiro et al. 2017; Meziadi et al. 2016; Chen et al. 2018) (Figure 1). Phylogenetically related NLs are spread between different chromosome ends, suggesting frequent sequence exchanges between non-homologous chromosomes. Our results highlight the importance of ectopic recombination in the evolution of R gene clusters and their distribution around the genome. This genome-wide analysis confirmed our previous analyses based on genome-wide khipu sequences or NL sequences from the Co-2 and B4 R clusters showing the plasticity of common bean subtelomeres (David et al. 2009; Richard et al. 2013; Chen et al.; 2018). This exceptional dynamic activity of subtelomeres has been reported in such diverse organisms as yeast and the malaria parasite Plasmodium (Brown et al., 2010, Current Biol.), but rarely in plants.
NL peculiar location, in proximity to heterochromatic regions (Figure 2), led us to study their DNA methylation status using a whole-genome cytosine methylation map (Collaboration with S.A. Jackson group; UGA; USA). In common bean, we showed that NL genes displayed an unusual DNA body methylation pattern since ~half of them are methylated in the three sequence contexts (CG, CHG, CHH), reminiscent of the DNA methylation pattern of repeated sequences (Richard et al. 2018a). Moreover 90 NLs were also abundantly targeted by 24 nt siRNA, with nearly all of them (95%) corresponding to methylated NL genes. This suggests the existence of a transcriptional gene silencing mechanism of NLs through the RdDM (RNA-directed DNA methylation) pathway in common bean which has not been described in other plant species (Richard et al. 2018). Consequently, in addition to the well described post-transcriptional gene silencing mechanisms of NLs involving microRNAs targeting NL mRNAs (Fei et al. 2016; MPMI), our results suggest the existence of an additional layer of regulation of NLs at the transcriptional level. Together, these mechanisms could be essential to down-regulate resistance expression in plant during normal growth condition, in absence of pathogen attack, to avoid the associated costs (Richard et al. 2018b). As expected, we have also identified in bean microRNAs targeting NL mRNAs and triggering the production of phased, secondary, small interfering RNAs (phasiRNAs) (Collaboration with Blake Meyers group; Danforth center; USA).
We are currently studying the dynamic of DNA methylation after pathogen infection at the genome wide level.
In addition to these genome wide analysis, we have a specific interest for anthracnose resistance clusters (Co-2, Co-x, B4) and for the virus resistance cluster (I cluster ) (Figure 1).
