InterATOME: Interactomic platform
Protein-protein interactions are critical elements of biological systems and their analysis can provide valuable hints about functions of proteins.
To date, the yeast 2 hybrid (Y2H) system is the only method to probe protein-protein interactions routinely in vivo and with a high-throughput. The Organelle Gene Expression-OGE team at IPS2 has been involved in an international project of Arabidopsis Interactome Mapping (1) coordinated by Pr Joseph Ecker (Salk Institute, San Diego) and Pr Marc Vidal (DFCI, Boston). This plant protein interactome network map was generated using the improved Y2H assay developed at DFCI for C. elegans, human and yeast proteins (2). The first data set from this project was published in 2011 (AIM, 2011) where a proteome-wide binary protein-protein interaction map containing about 6200 high quality interactions between about 2700 proteins was described. The AIM-dataset is totally accessible to the scientific community : Plant Interactome Database .
The OGE team fully automated and improved the Y2H protocol which was previously used for the AIM-project. Using 384-well plates, successive culture replication in liquid media and robots, the SPOmics-Interactomic Platform is able to screen a pool of 50 DB-X hybrid proteins against the AIM AD-library (about 12000 Arabidopsis proteins) within two months.
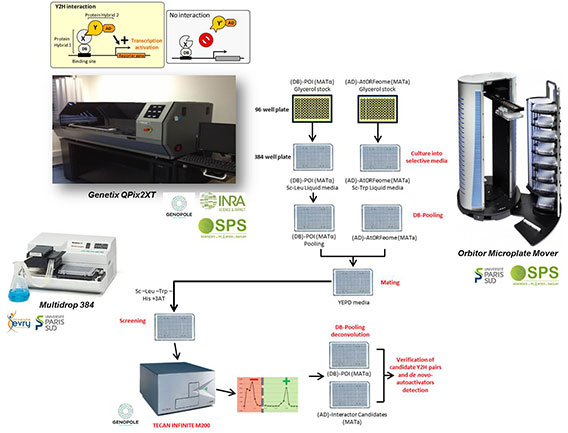
Briefly, ORFs-baits provided by our collaborators directly into the pDEST-DB vector previously supplied from the platform, are utilized for yeast transformation and DB-X hybrid proteins are tested for autoactivator phenotype. DB-X hybrid protein expressing yeasts are then individually grown in selective media, combined into pools of 50 clones and screened against the AIM AD-library by mating. After identification of candidate interacting AD-proteins, a matricial Y2H assay is performed by de-pooling of the 50 DB-preys and testing against individual candidate AD-baits. A final sequencing step allows unambiguous identifications of both AD- and DB- interacting proteins.
Every new screened ORF is integrated into the AD-library allowing a continuous network implementation.
Cost (academic lab): 12,500 euros for a screening of up to 50 DB-X hybrid proteins.
(1) Evidence for Network Evolution in an Arabidopsis Interactome Map. Arabidopsis Interactome Mapping Consortium. Science 29 Jul 2011: Vol. 333, Issue 6042, pp. 601-607
(2) High-Quality Binary Interactome Mapping. Dreze M, Monachello D, Lurin C, Cusick ME, Hill DE, Vidal M, Braun P. Methods in enzymology 470:281-315 January 2010
