Research
Happy New Year 2026
Read moreResearch
Christmas at IPS2
Read moreResearch
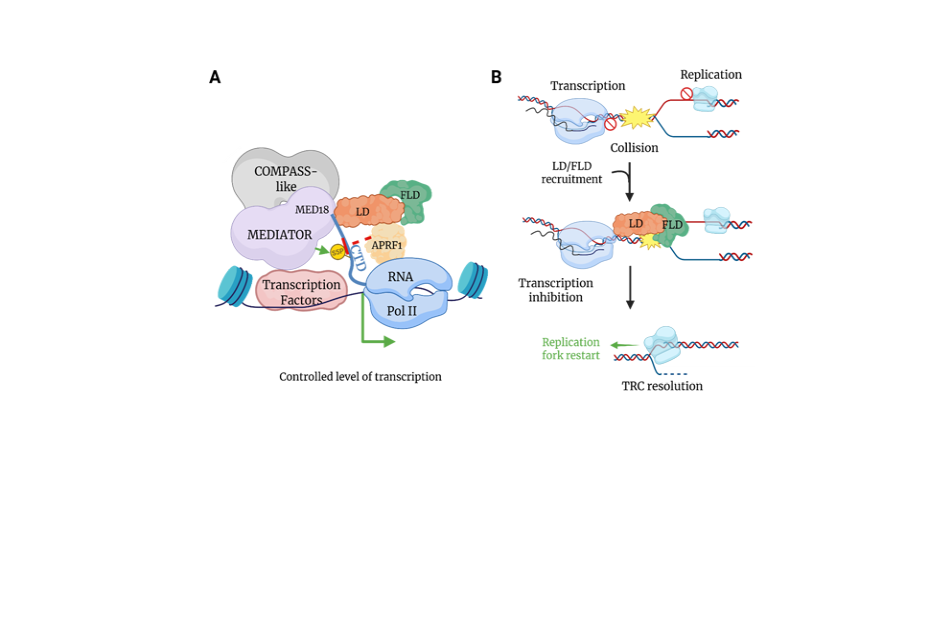
Global transcription repression in plants
Read moreResearch
William BOUARD has joined the CCARS team
Read moreResearch
A new ERC project at IPS2!
Read moreResearch
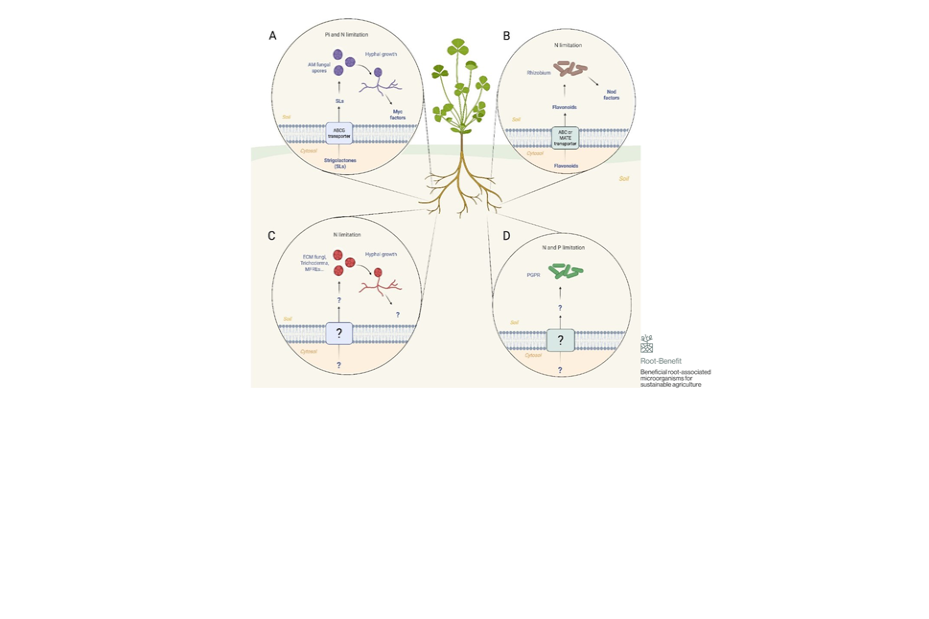
A review on molecular mechanisms of beneficial plant-microbe interactions
Read moreResearch
Saclay Plant Sciences (SPS) Summer School 2026
Read moreResearch
Launch of the HeatDDR Doctoral Network
Read moreResearch
High-throughput and low-cost transcriptomics
Read moreResearch
Arrival of Bruno Guillotin at IPS2
Read moreResearch
A wind of music is blowing through our institute!
Read moreResearch
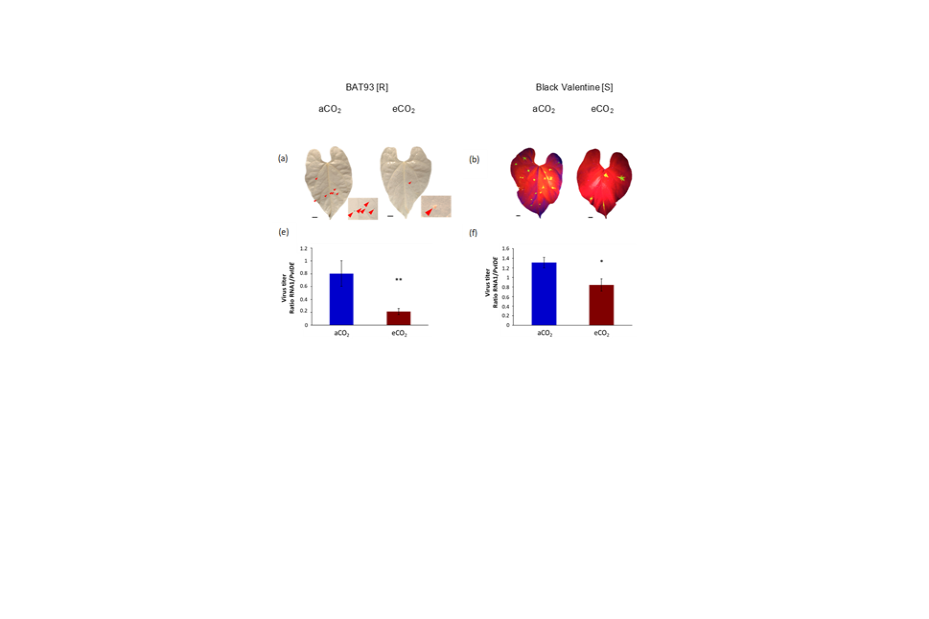
Elevated CO2 enhances resistance to the BPMV virus in common bean
Read moreResearch
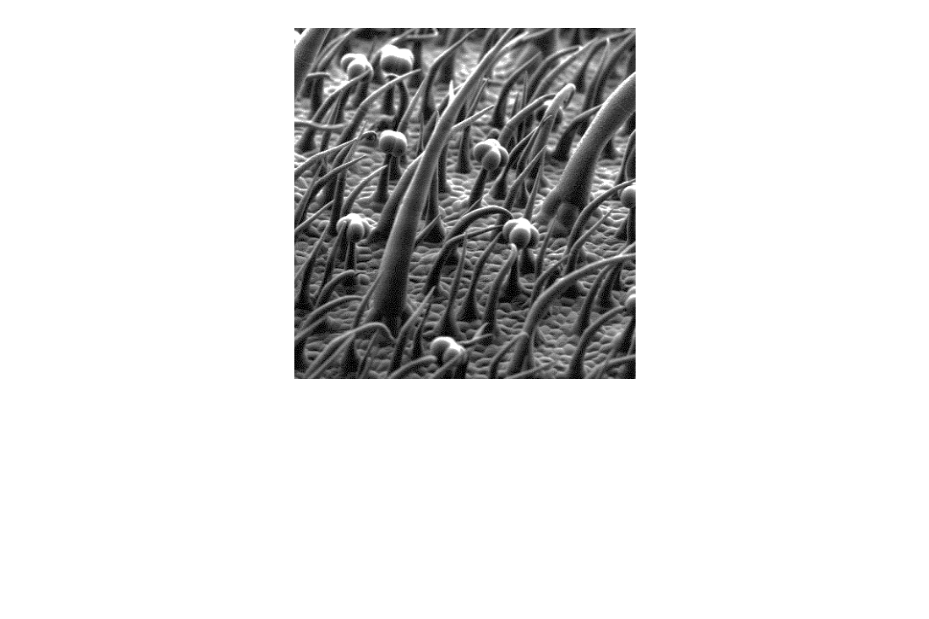
EXPLOR’AE PROJECT YETI “Unravelling the secrets of trichomes”
Read more