Regulatory non-coding RNAs in root plasticity
Team REGARN / Martin Crespi
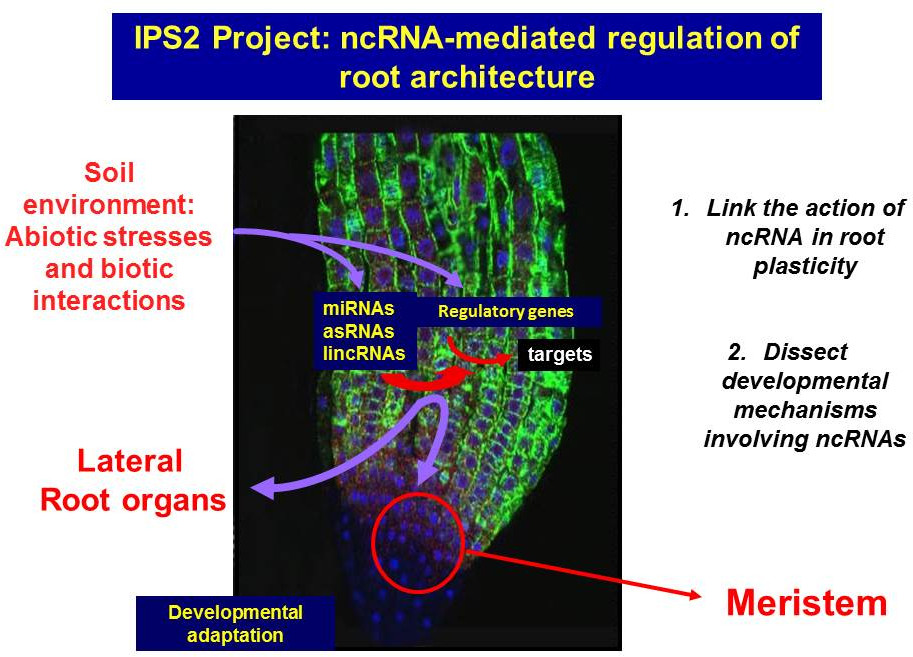
Scientific objectives and strategy
Genomic approaches changed our view of the eukaryotic transcriptome. One of the major discoveries linked to these large scale sequencing was that the number of non-protein-coding transcripts, generally called non-coding RNAs (or ncRNAs) is much larger than previously believed, including microRNAs, small-interfering siRNAs and high numbers of long ncRNAs or lncRNAs (intergenic or antisense, Ariel et al., 2015). In the last years, many lncRNAs were shown to act as positive or negative quantitative regulators of gene expression. Variation in the nucleotide sequences or expression patterns of the non-coding genome can have less pleiotropic effects than changes in the protein sequence of critical regulators. Therefore, in addition to promoters, introns and transposons, regions encompassing non-coding RNAs emerge as actors of plant adaptation to environmental constraints.
M. Crespi / REGARN team
Developmental mechanisms involving ncRNAs are under active investigation. Both miRNAs and siRNAs play essential roles in eukaryotes by guiding target mRNA cleavage or translational repression after integration into a ribonucleoprotein complex, the RISC (RNA Silencing Induced Complex). Indeed, miRNAs and certain siRNAs participate in complex regulatory loops with their targets (generally transcription factors) to control several aspects of root development (e.g. Lelandais-Brière et al., 2016). In contrast, the action of lncRNAs and the majority of the siRNAs is much less known (Ariel et al., 2015). Root development and growth are complex processes integrating a large variety of signalling pathways (endogenous such as hormones or exogenous such as nutrients or soil water content) resulting in different root architectures. Thus, the root is able to adjust its growth and development in response to environmental cues. Modifying spatially or temporally the action of miRNAs or other regulatory lncRNAs is one way for plants to integrate signals from the environment into future growth and development. Another model of root plasticity is the ability of legume plants to interact with bacterial symbionts to develop new organs, the nitrogen-fixing nodules. Root nodules are novel organs where expression reprogramming of thousands of genes occurs in both symbionts, the bacteria and the plant host cell, in a defined developmental context. Hence, it is a nice model for understanding the role of regulatory RNAs in organogenesis.
Our project aims to understand the role of ncRNAs in root plasticity. At IPS2, our lab develops two research axes using Arabidopsis thaliana and Medicago truncatula to address key emerging questions on the non-coding RNA field:
- To link ncRNAs to root plasticity by exploring genome and ecotype diversity
- To dissect developmental mechanisms triggered by ncRNAs in root developmental processes