Organellar gene expression
Our main focus is to understand the regulation of gene expression in organelles.
Comprehending the intricate functions and regulatory mechanisms within plant organelles is fundamental to advancing crop productivity and enhancing their resilience to variable environmental conditions. Plastids and mitochondria, the primary sites of energy metabolism in eukaryotic cells, also wield substantial influence over various metabolic pathways. Over the course of evolution, these organelles have undergone substantial genome reduction through the process of endosymbiotic events, leading to the transfer of genetic material to the cell nucleus. Consequently, the metabolic processes within organelles depend heavily on proteins encoded by the nucleus and subsequently targeted to these organelles. Nevertheless, these organelles have retained compact genomes that house essential proteins and RNAs pivotal for their biological functions, necessitating a coordinated control of nuclear and organellar gene expressions. Various processes governing organellar transcription and post-transcriptional activities, including DNA replication, RNA transcription, RNA processing, RNA editing, RNA splicing, and translation, rely on proteins encoded in the cell nucleus. Within this spectrum of nuclear factors, the expansive family of pentatricopeptide repeat proteins (PPR) stands as a pivotal element in the orchestration of organellar genome regulation. These nuclear-encoded proteins are almost exclusively directed to mitochondria or chloroplasts, where they specifically bind to RNA molecules, actively participating in all facets of gene expression, encompassing transcription through translation. Complementing this regulatory role are ribonucleases, both endo- and exo-ribonucleases, which contribute to the processing of nascent transcripts, culminating in the formation of the mature organellar transcriptome. This complex network of interactions (protein-RNA, protein-protein, DNA-protein) is dynamic and occurs mainly in the nucleoid, a nucleo-protein complex still poorly characterized.
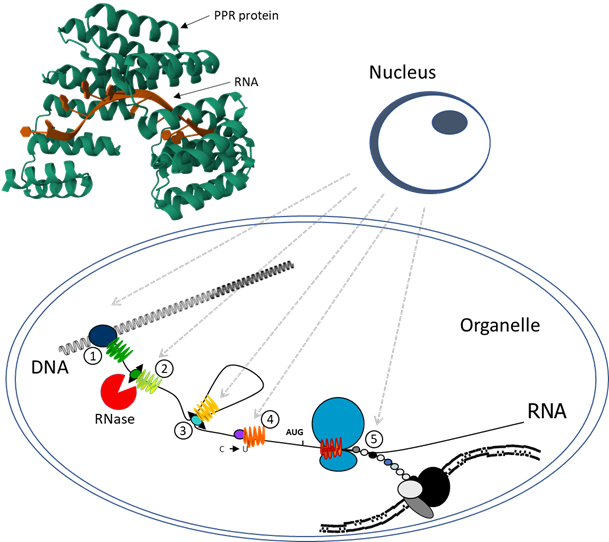
[“Roles for PPR proteins and ribonucleases in organellar gene expression: from transcription (1) to translation (5). (2) RNA cleavage and end maturation, (3) splicing, (4) editing. “]
Historically, our team played a pioneering role in the discovery of the PPR family back in 2000. Subsequently, we conducted genomic characterizations of this family in Arabidopsis, rice, and Physcomitrium. We have also undertaken various genetic and functional studies on members of the Arabidopsis family. Over time, our research focus has broadened to encompass plastid ribonucleases and the investigation of organellar gene expression in dynamic contexts, such as during development or under stress conditions, as part of three principal projects:
In parallel, the OGE team develops functional genomics tools available for the plant community through the activity of two technological platforms: the Paris-Saclay Plant Transcriptomic Platform and the SPOmics Interactome Platform.
The SPOmics Interactome Platform originated from the active participation of the OGE team in an international project focused on Arabidopsis Interactome Mapping. This collaborative effort was coordinated by Professors Joseph Ecker (Salk Institute, San Diego) and Marc Vidal (DFCI, Boston). The plant protein interactome network map was constructed using an enhanced Y2H assay originally developed at DFCI for studying protein interactions in C. elegans, human, and yeast proteins. The initial dataset from this project was published in 2011 (AIM, 2011). It featured a comprehensive binary protein-protein interaction map, encompassing approximately 6200 high quality interactions between about 2700 proteins was described.
