Organelles at the core of plant stress responses
Plastids are the site of photosynthesis and of various metabolic reactions, including amino acid, lipid, starch, and sulfur metabolism that are all strongly affected by environmental variations. Moreover, several pathogens target plastids with effectors as part of their virulence strategy and plastids are involved in the synthesis of key phyto-hormones involved in plant pathogen interactions. They are also a major source of reactive oxygen species involved in Pathogen- and Effector-triggered immunities. Thus, being central to photosynthesis and primary metabolism, plastids integrate, decode and respond to cellular and environmental signals including biotic and abiotic stresses. our results also point towards a major role of chloroplast in stress response. Using our SPOMics-InterATOME platform, we have identified that several plastid-located proteins are targeted by fungal and bacterial pathogen effectors and that a single stress related cytoplasmic protein interacts with all plastid encoded proteins. Furthermore, the stress resilience gene network of Arabidopsis that we identified through a meta-analysis, in close collaboration with the Genomic Networks team, of the data produced by the POPS platform supports the importance of plastid gene expression in stress response. A detailed analysis of these networks provides a number of interesting candidates which we are currently characterizing.
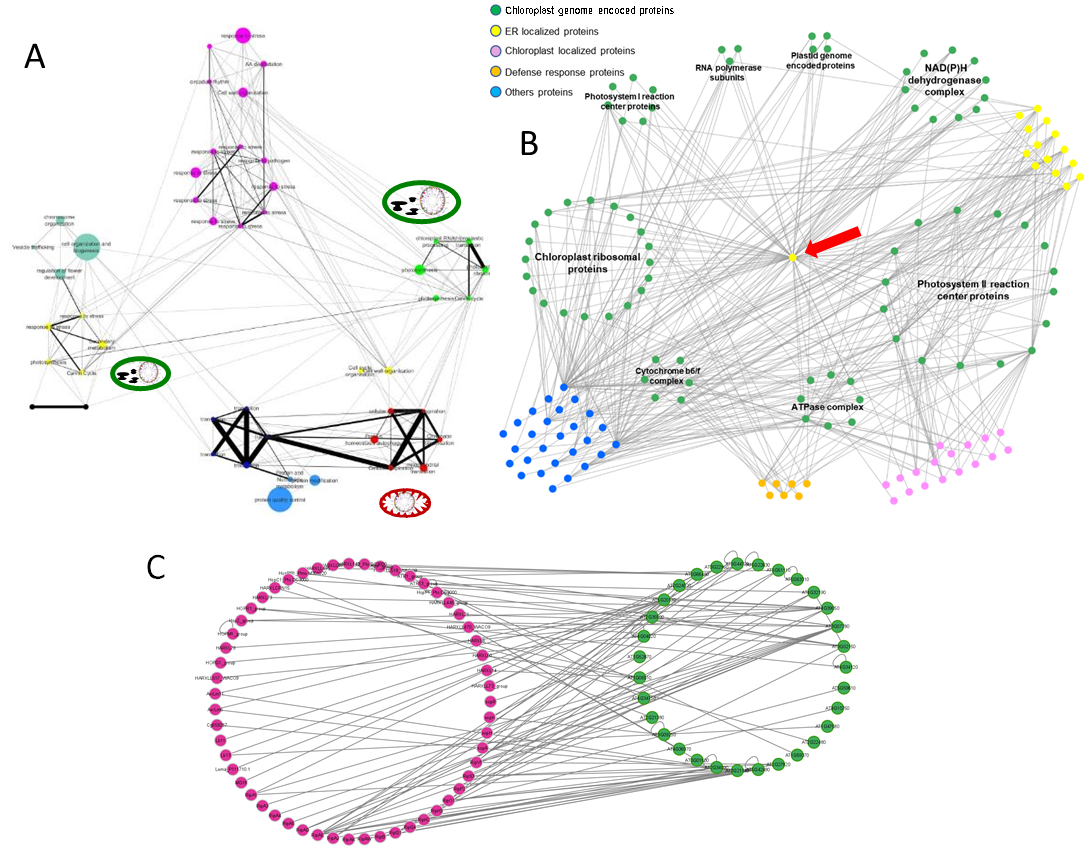
Figure: plastids at the core of plant stress response. A: the stress resilience network of Arabidopsis thaliana contains numerous gene clusters associated with plastid functions (green). B: The plastid encoded proteins (green dots) are interacting in high-throughput yeast-2-hybrid with several defense- (orange dots) and ER-related (yellow dots) proteins including one ER-related protein interacting with all of them (red arrow). C: Several pathogen effectors (purple dots) are interacting with proteins targeted to plastids (green dots) in high-throughput yeast-2-hybrid.
