Positional Cloning
The main focus of this activity is the valorisation of the genetic material created in INRAE. This is carried out through cloning and characterisation of genes of agronomic importance used or susceptible to be used in breeding. Three distinct research areas are at the core of the positional cloning activities: (i) establishment of state of the art tools for positional cloning (ii) enquiring for genes of agronomic importance and (iii) establishment of collaborations required for cloning and characterisation of the identified genes. The target phenotypes could emerge from phenotyping of germplasms or from EMS mutagenized collections. For instance, several mutant collections have been phenotyped and the data made available for the scientific community by UTILLdb database (Dalmais et al., 2008). This far UTILLdb manage phenotypic data of pea, Brachypodium and flax. This resource has an open access and all seed lines of interest can be ordered via our contacts.
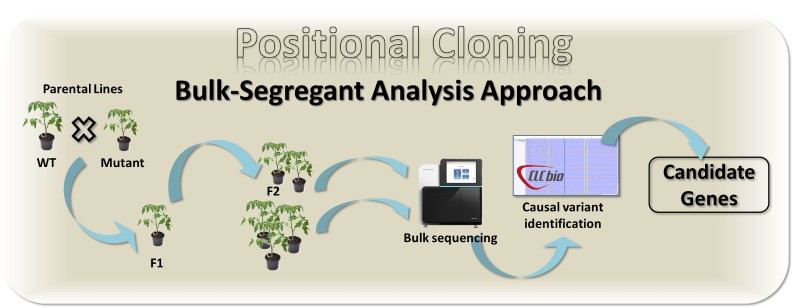
Our positional cloning workflow is schematized below.
EPITRANS offers its expertise at different levels, potentially combined, depending on objectives, materials and resources:
- Screening a chemically induced mutant collection for a given phenotypic trait.
- Sequencing segregant pools followed by bio-informatics analysis to identify the causal mutation
- Mapping and functional validation of the identified candidate genes.
As example we isolated (i) genes conferring resistance to aphids (Vat), viruses (nsv, pvr2, ZYM, etc.), fungi (Fo2, PmW, etc), and bacteria (Bs), (ii) Sex determination genes (M, G, A, etc), (iii) male fertility restoration gene (Rfo), and flowering and development genes (Ti, Rms2, HR, J-2, etc). Eight patents have been registered and over 60 articles published.
