Hormonal crosstalks regulating symbiotic nodulation and root development
Team SILEG / Florian Frugier
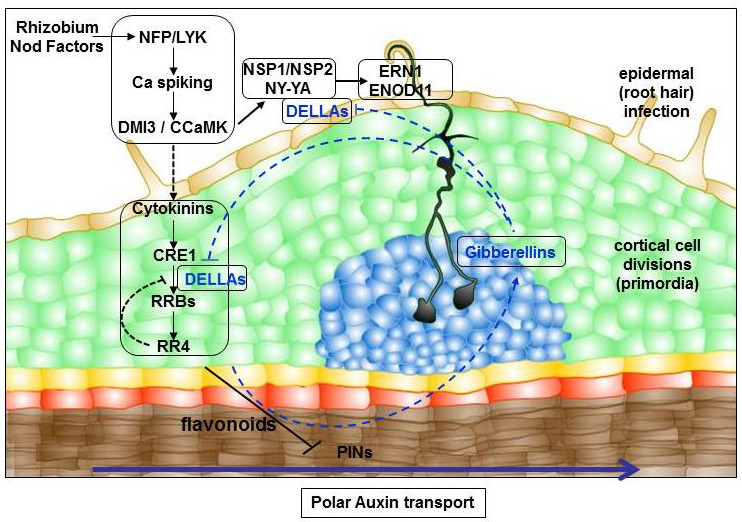
We showed that cytokinin phytohormones regulate antagonistically symbiotic nodule and lateral root development depending on the CRE1 (Cytokinin Response 1) receptor (Gonzalez-Rizzo et al.,2006 ; Frugier et al., 2008 ; Plet et al., 2011 ; Laffont et al., 2015). Even though rhizobia bacteria synthesize and secrete typical bio-active cytokinins, their contribution to nodulation seems nevertheless rather limited (Kisiala et al., 2013). The CRE1 cytokinin signaling pathway regulates nodule organogenesis through the modulation of polar auxin transport (Plet et al., 2011) depending on the accumulation of specific flavonoids (Ng et al., 2015), leading to a local auxin accumulation in dividing cortical cells associated to early nodule formation. CRE1 acts redundantly with other CHASE Histidine Kinase (CHK) cytokinin receptors in nodule initiation, and can be replaced by the closest homolog from the Arabidopsis thaliana aposymbiotic plant (Boivin et al., 2016). Cytokinins and CHK receptors have divergent functions in mature nodules, notably in relation to the nitrogen fixation capacity, which can not be replaced by the Arabidopsis CRE1 gene. In addition, cytokinins and the CRE1 signaling pathway also act in the root epidermis where they seem to negatively regulate bacterial Nod factor signaling and potentially rhizobial infections (Jardinaud et al., 2016).
A combination of biochemical and transcriptomic analyses allowed us to identify in M. truncatula roots primary cytokinin response genes depending on the CRE1 signaling pathway (Ariel et al., 2012). Among those, the Nodulation Signaling Pathway 2 (NSP2) transcription factor, previously demonstrated to be critical for nodule organogenesis, was identified. Interestingly, this gene is also posttranscriptionally regulated in response to cytokinins by a specific microRNA isoform (miR171h). This strategy also led us to identify a new basic Helix-Loop-Helix (bHLH) transcription factor (MtbHLH476) as a cytokinin primary response gene positively involved in nodule formation. More recently, we showed that a specific type B Response Regulator (RRB) transcription factor, MtRRB3, regulates nodulation and targets the promoters of NPS2 and of an endoreduplication cycle progression regulator, MtCCS52A (Cell Cycle Switch 52A; Tan et al., 2020).
These transcriptomic analyses also allowed to identify that the gibberellin (GA) phytohormone metabolism was rapidly regulated by cytokinins. We then showed that GA indeed negatively regulates nodulation, and notably rhizobial infections, depending on DELLA signaling proteins (Fonouni-Farde et al., 2016). Strikingly, DELLA proteins in the epidermis are sufficient to induce a symbiotic infection marker, thanks to the interaction with the critical Nod Factor signalling components NSP2 and NF-YA1 to transactivate ERN1 gene expression. In addition, expression of a DELLA protein in the cortex induces nodule-like structures and allows to complement the cre1 mutant nodulation defective phenotype, indicating that DELLA proteins also act downstream of the CRE1-dependent cytokinin pathway (Fonouni-Farde et al., 2017). Accordingly with a gibberellin/cytokinin crosstalk, GA negatively regulates the cytokinin pool and response, as well as the cytokinin regulation of NSP2 and ERN1 early nodulation genes. GAs also regulate the development of the root system, including the number of cortex layers (Fonouni-Farde et al, 2019), and these pleiotropic effects do not allow to safely conclude about the role of these hormones in nodule organogenesis.
In addition to the role of the CRE1 cytokinin signaling pathway in symbiotic nodulation, we analyzed its involvement in other root environmental responses. Whereas no defect in endomycorhization was observed in cre1 roots, an increased tolerance to an abiotic stress (salt) and to root pathogens (Ralstonia solanacearum and Aphanomyces euteiches) was identified (Moreau et al., 2014 ; Laffont et al., 2015). In addition to direct cross-talks between cytokinin and defense/stress responses, these phenotypes additionally correlate with the increased ability of the cre1 mutant to form lateral roots.
