Stress responses and nodule senescence
Team SILEG / Florian Frugier
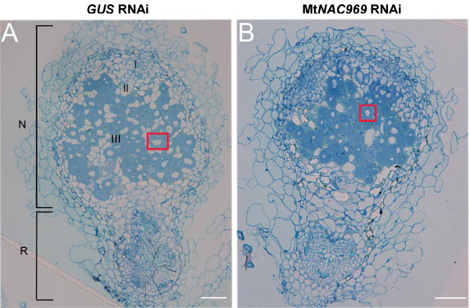
We characterized across different studies the adaptive root architecture responses induced by an abiotic stress (salt) in M. truncatula and identified thanks to various transcriptomic approaches different markers and regulatory genes regulated in response to salt stress and linked to root and nodule development (Frugier et al., 2000 ; Gargantini et al., 2006 ; de Lorenzo et al., 2007 ; Chinchilla et al., 2008 ; Gruber et al., 2009 ; Zahaf et al., 2012). Among those, a connection with cytokinin signaling was highlighted (Merchan et al., 2007 ; Laffont et al., 2015). In addition, we characterized a transcriptional network involving a type I HomeoDomain-Leucine Zipper (HD-ZIP) transcription factor regulating a LOB (Lateral Organ Boundary) transcription factor, which controlled lateral root emergence notably under abiotic stress conditions (Ariel et al., 2010). Finally, we identified a salt-regulated NAC transcription factor that is involved both in root response to an abiotic stress and in symbiotic nodule senescence (de Zélicourt et al., 2012). More recently, to identify nodule Senescence Associated Genes (SAGs), transcriptomic analyses of aging developmentally senescent nodules, of the basal region of nodules containing the senescence zone, and of nitrate versus salt treated nodules (environmentally-induced senescence) were performed (Sauviac et al., 2022). Among the 243 genes at the intersection of these four senescence-inducing conditions were the previously identified NAC gene, validating the strategy used. In addition, among hormonal related genes, many cytokinin regulated genes were identified, and increasing the cytokinin content specifically in the nodule senescence zone (using a pCP6:IPT construct) promoted the nitrogen fixation / senescence transition, unlike in leaves where cytokinins delay senescence.