In depth transcriptomics of symbiotic nodule senescence
A dual legume-rhizobium transcriptome of symbiotic nodule senescence reveals coordinated plant and bacterial responses
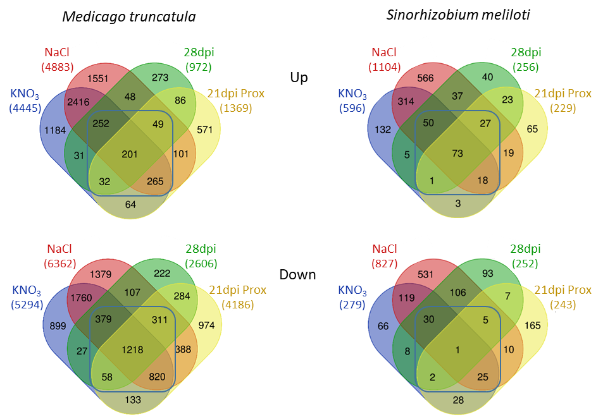
Senescence determines plant organ lifespan depending on aging and environmental cues. During the endosymbiotic interaction with rhizobia, legume plants develop a specific organ, the root nodule, which houses nitrogen (N)-fixing bacteria. Unlike earlier processes of the legume–rhizobium interaction (nodule formation, N fixation), mechanisms controlling nodule senescence remain poorly understood. In this collaborative work involving the SILEG team at IPS2 and published in Plant Cell & Environment, in order to identify nodule senescence-associated genes, we performed a dual plant-bacteria RNA sequencing approach on Medicago truncatula-Sinorhizobium meliloti nodules having initiated senescence either naturally (aging) or following an environmental trigger (nitrate treatment or salt stress). The resulting data allowed the identification of hundreds of plant and bacterial genes differentially regulated during nodule senescence, thus providing an unprecedented comprehensive resource of new candidate genes associated with this process. Remarkably, several plant and bacterial genes related to the cell cycle and stress responses were regulated in senescent nodules, including the rhizobial RpoE2-dependent general stress response. Analysis of selected core nodule senescence plant genes allowed showing that MtNAC969 and MtS40, both homologous to leaf senescence-associated genes, negatively regulate the transition between N fixation and senescence. In contrast, overexpression of a gene involved in the biosynthesis of cytokinins, well-known negative regulators of leaf senescence, may promote the transition from N fixation to senescence in nodules.
05/09/2022
