How the genome “dark matter” regulate distant genes ?
A long non-coding RNA affects 3D chromatin conformation of distant loci through DNA-RNA hybrids or R-loops
In the last years, a large portion of the eukaryotic transcriptome which does not code for any functional protein, the so-called non-coding RNAs, have emerged as regulators controlling gene expression. This “dark matter” of the eukaryotic genome, albeit not translated into proteins, can be transcribed in functional noncoding RNAs participating in a wide range of molecular mechanisms throughout evolution. In previous work, the team of Martin Crespi at the Institute of Plant Sciences Paris Saclay (IPS2) revealed that the APOLO long non-coding RNA was able to control the expression of a key regulator of plant hormone signaling (the PID gene) in cis through modulation of PID promoter accessibility (Ariel et al., Molecular Cell 2014). This year, in a work also published in Molecular Cell (online 14th of January), the team from IPS2 in collaboration with colleagues from the Institute of “Agrobiotechnology from the Litoral” (IAL, Argentina), in the frame of a NOCOSYM LIA CNRS (“Laboratoire International Associé”) demonstrated that the APOLO long noncoding RNA can coordinate expression of multiple distant genes in trans in Arabidopsis. APOLO recognizes these targets through sequence complementarity and the formation of DNA-RNA duplexes, named R-loops.
Using several genome-wide methodologies, the authors could show that, upon target recognition, APOLO can modulate the three-dimensional chromatin conformation of the distant loci to fine-tune their transcription. This R-loop-dependent recognition can decoy key epigenetic regulators (such as LHP1, a protein involved in Polycomb-related complexes) from these distant sites in order to modulate their chromatin accessibility. Thus, the activity of the noncoding genome is dynamically shaping the spatial organization of genetic information in the nucleus at several loci.
This study revealed a new regulatory mechanism involving non-coding RNAs that may contribute to the wide diversity of gene expression patterns observed in specific cell types in eukaryotes. Hence, in the “genome universe”, the genetic “dark matter” (the non-coding RNAs) may regulate the “brightness” (expression levels) of the “stars” (protein coding genes).
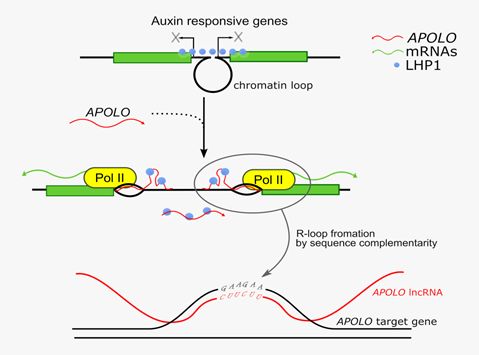
Trans-action of APOLO long noncoding RNA over a plethora of auxin-responsive genes.
