Splicing regulation by long noncoding RNAs
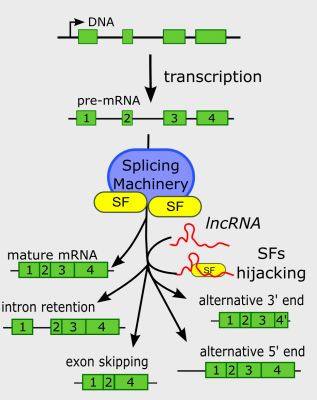
In the last years, the advent of novel sequencing techniques allowed the identification of thousands of RNAs that are expressed all along the genome that do not seem to code for proteins as usual mRNAs do (except few well characterized cases such as ribosomal and transfer RNAs). These noncoding RNAs, a kind of “dark matter” of the genome, have been linked to a wide range of regulatory mechanisms controlling protein-coding mRNA expression such as mRNA stability, translational repression or interactions with the transcriptional and chromatin machineries. Recently, a new link between noncoding RNA and mRNA splicing is emerging. Splicing is the process by which introns, noncoding segments of an mRNA, are removed from the initial transcript in order to obtain a mature mRNA able to be translated into a protein. In certain circumstances, splicing reactions can be modulated generating two or more mRNA isoforms from a single pre-mRNA by a process called alternative splicing. Then, alternative splicing can boost the plasticity of the eukaryotic proteome. In a recent review, Romero-Barrios and colleagues from IPS2 at Paris Saclay and IAL in Argentina, discuss recent discoveries about the different mechanisms by which long and small noncoding RNAs modulate alternative splicing. These analyses allowed them to classify these mechanisms according to the commonalities and singularities of their interaction with the splicing machineries, their direct association with pre-mRNAs, their roles in nuclear localization processes or their impact on chromatin conformation and epigenetic regulation of transcription. Hence, noncoding RNAs may fine-tune the splicing process to expand mRNA and consequently protein diversity in living organisms and can define novel biotechnological strategies to regulate gene expression in eukaryotes.