Mitochondrial translation in plants is initiated by a protein-RNA interaction
Progress in plant mitochondrial translation
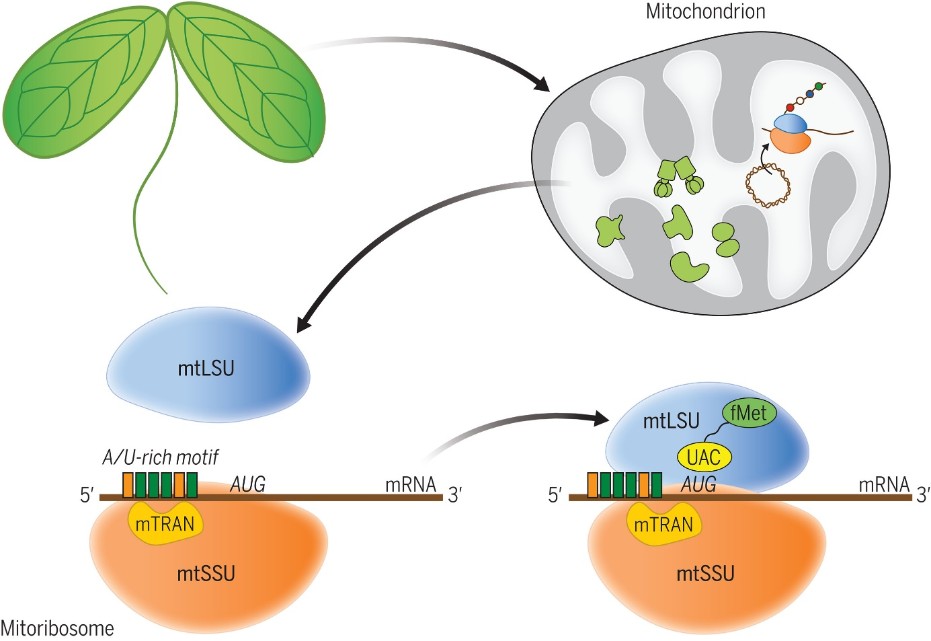
Mitochondria, involved in the energy metabolism of eukaryotic cells, derive from the endosymbiosis of a bacterial ancestor by a primitive eukaryotic cell. Although all mitochondria derive from the same common ancestor, the protein complexes essential to their function have subsequently diverged widely between the different eukaryotic groups. This is particularly true of the ribosome, the complex that translates messenger RNA into proteins. In plant mitochondria, the mechanism by which translation is initiated remained a mystery. In bacteria, initiation depends on the ribosome attaching a specific mRNA sequence (called Shine-Dalgarno), which is absent in the messenger RNAs of plant mitochondria. In a study published in the journal Science, a consortium of researchers led by Olivier van Aken from Lund University (Sweden) and including the 'Organelle and Reproduction' team at the Institut Jean-Pierre Bourgin and the 'Genomic Expression of Organelles' and 'Genomic Networks' teams at the Institut de Science des Plantes Paris-Saclay, has identified an essential role for mTRAN proteins in translation initiation in the mitochondria of the Arabidopsis thaliana plant. The mTRAN proteins are conserved in all land plants, form part of the mitochondrial ribosome and are capable of binding to highly conserved A/U-rich motifs in mRNAs. The mTRANs could thus enable translation initiation by acting as a guide for the ribosomes thanks to an RNA-protein interaction mechanism that differs from that of bacteria or mammalian mitochondria.