A long noncoding RNA modulates the transcriptome by interacting with multiple core splicing factors
Alternative splicing (AS) is a major source of transcriptome diversity. Long noncoding RNAs (lncRNAs) have recently emerged as regulators of AS through different molecular mechanisms. In Arabidopsis thaliana, the AS regulators Nuclear Speckle RNA binding proteins (NSRs) interact with the ALTERNATIVE SPLICING COMPETITOR (ASCO) lncRNA. In a study published in EMBO reports (Rigo et al., 2020), the REGARN team from IPS2 analyzed the effect of the knock‐down and overexpression of ASCO at the genome‐wide level and found a large number of deregulated and differentially spliced genes related to plant defense responses. In agreement, ASCO-silenced plants are more sensitive to flagellin, a molecule that mimics the attack by a pathogen. Using biotin‐labeled oligonucleotides for RNA‐mediated ribonucleoprotein purification, the team showed that ASCO binds to the highly conserved spliceosome component Pre-mRNA PROCESSING 8a (PRP8a). Moreover, ASCO overaccumulation impairs the recognition of PRP8a transcript targets, suggesting that ASCO can directly bind to the core of the spliceosome. They further showed that ASCO also binds to another spliceosome component, the Small nuclear ribonucleoprotein Sm D1b (SmD1b). This indicates that, besides NSRs, ASCO interacts with multiple core splicing factors in the same way as small nuclear RNAs U1 to U6. Hence, lncRNAs may integrate a dynamic network including conserved spliceosome core proteins. How sequence-divergent lncRNAs can interact with highly conserved components of the spliceosome machinery yet remains a major question and points towards new mechanisms by which lncRNAs could modulate transcriptome reprogramming in eukaryotes.
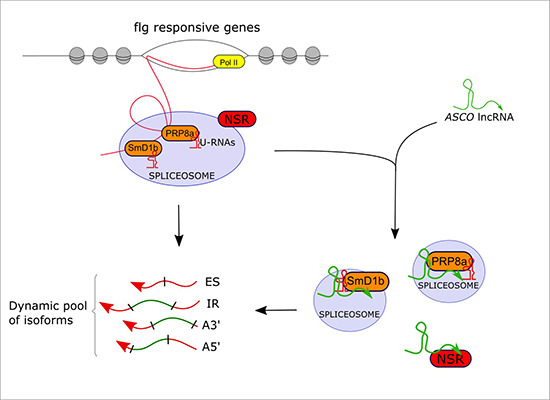
Schematic model of ASCO’s lncRNA role in the modulation of alternative splicing of specific transcripts.
The lncRNA ASCO interacts with the highly conserved splicing factors PRP8a and SmD1b to modulate the alternative splicing of flagellin responsive transcripts.
