An unexpected CLE peptide regulates symbiotic nodulation of legume plants
An improved linkage disequilibrium-based statistical test used for Genome-Wide Epistatic Selection Scans allowed predicting a genetic interaction between a new CLE signaling peptide and the SUNN receptor
The quest for signatures of selection using single nucleotide polymorphism (SNP) data has proven efficient to uncover genes involved in conserved and/or adaptive molecular functions, but none of the statistical methods were designed to identify interacting alleles as targets of selective processes. In a collaborative study between the Frugier team at IPS2 and M. Bonhomme at the LRSV (Toulouse, France), published in Heredity, we propose a statistical test aimed at detecting epistatic selection, based on a linkage disequilibrium (LD) measure accounting for population structure and heterogeneous relatedness between individuals. SNP-based (Trv) and window-based (TcorPC1v) statistics both fit a Student distribution, allowing testing the significance of correlation coefficients. The use of this improved statistical test on SNP data from the Medicago truncatula symbiotic legume plant allowed to uncover a previously unknown gene coadaptation between the MtSUNN (Super Numeric Nodule) receptor and the MtCLE02 (CLAVATA3-Like 02) signaling peptide. Experimental evidence allowed to demonstrate as a proof of concept that this coadaptation signature was associated to a genetic link between these two genes, as the MtCLE02 signaling peptides regulate negatively symbiotic root nodulation depending on the MtSUNN receptor. This collaborative work was made possible thanks to the ANR project “PSYCHE” coordinated by F. Frugier at IPS2.
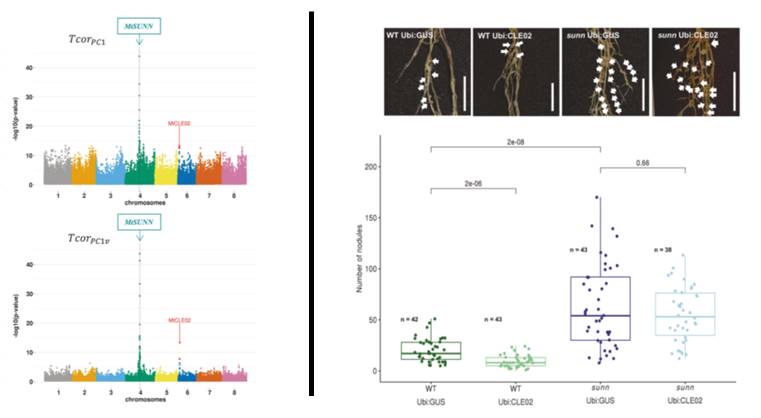
Figure. From the identification of a coadaptation signature between two Medicago truncatula genes to the experimental evidence of their genetic interaction.
On the left, the LD distribution calculated from TcorPC1 (above panel) and from
TcorPC1v statistics (lower panel) between the bait gene MtSUNN (framed) and all genes of the M. truncatula genome. The x-axis corresponds to gene positions spanning the eight chromosomes, each point corresponding to a gene and the MtCLE02 gene being indicated with an arrow. The y-axis shows the -log10(p value) of the test of the correlation coefficient. On the right, number of symbiotic nodules formed on control roots (Ubi:GUS) or roots overexpressing the MtCLE02 signaling peptide (Ubi:CLE02), in Wild-Type plants (WT; two left images and bars) and in a mutant of the SUNN receptor (sunn; two right images and bars). This demonstrates that the negative effect of CLE02 on nodule numbers requires the SUNN receptor activity.
