Spatial genome organization and gene co-regulation in Arabidopsis
The regulation of gene expression during cell differentiation in eukaryotes is closely linked to covalent modifications of chromatin. Among these, post-translational modifications of histones influence the level of chromatin compaction, thus modulating the accessibility of different regions of the genome to the transcription machinery. Since a few years, the concept of genetic information organized in a linear way as a sequence of nucleotides associated with histones has evolved towards a more complex view of this structure, integrating the dynamics of the three-dimensional architecture of the nucleus. The development of nuclear interactome analysis methods has revealed the existence of key architectural elements and their major functional role in the regulation of physical interactions between promoters and regulatory intergenic elements or between sets of genes.
In a study published in Genome Research, the Chromosome Dynamics team (IPS2, CNRS, Université Paris-Saclay, INRAE) investigated the role of post-translational modification of histone H3K27me3 in the control of chromatin architecture and spatial organization of genes targeted by Polycomb protein complexes in Arabidopsis thaliana. By combining cytology, genetics, transcriptomics, epigenomics and 3D analysis of chromatin conformation, they were able to show that H3K27 methylation defects lead to a reconfiguration of chromatin architecture through a reorganization of contacts between genes. This work illustrates the role of histone modifications on the formation of chromatin interactions, a key element of gene co-regulation during plant development.
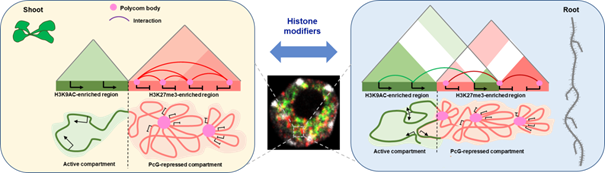
Figure legend: Histone modification complexes control chromatin architecture by inducing the formation of repressive and active domains to enable co-regulation of genes in specific tissues of Arabidopsis seedlings. The green and pink structures represent respectively the active compartments associated with the H3K9ac chromatin modification and the compartments repressed by Polycomb group (PcG) proteins, associated with the H3K27me3 modification.
