Low complexity drives transcriptional regulatory rules
Minimum complexity drives regulatory logic in Boolean models of living systems
Cellular decisions depend on the coordinated action of molecular components. Their modeling can be tackled via Boolean networks, a mathematical framework where directed edges represent dependences and where a logic rule specifies how a gene’s state is determined by the values of its regulatory inputs. The construction of Boolean regulatory models requires a substantial amount of experimental data on the interactions between the underlying network components. Though the structure of such networks has been studied quite extensively, a comprehensive characterization of the logic rules has not been carried out so far.
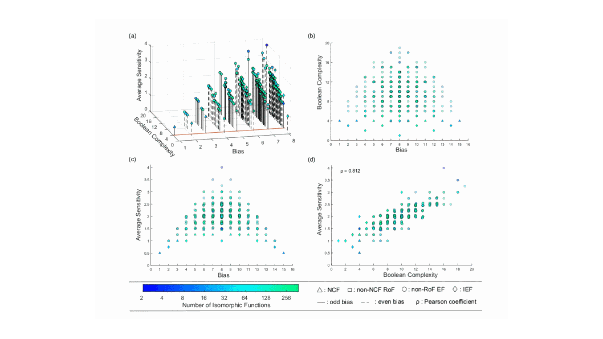
In a study published in PNAS Nexus, an IPS2 researcher (Team REGARN) and a team at IMSc, Chennai (India), compile 2687 logic rules from 88 reconstructed discrete models reported in the Boolean modeling literature. First, they find that these rules obey biologically meaningful properties such as ‘effectiveness’, ‘unateness’ and ‘canalyzation’. Second, they find that two types of rules are highly over-represented, namely: read-once functions and nested canalyzing functions. Interestingly, these two types have the lowest complexity based on two measures: Boolean complexity (based on the string lengths defining the logic rule) and average sensitivity. This analysis indicates that simple rules are preferentially used in complex cellular decisions.
19/05/2022
