The “Chloroplast protein-protein interaction” project
The InterATOME platform (IPS2) has been recently involved in the international Arabidopsis Interactome Mapping - AIM project to establish the first map of the plant protein interaction network, as well as in the construction of the largest database of pathogen effector interactors through several international collaborations.
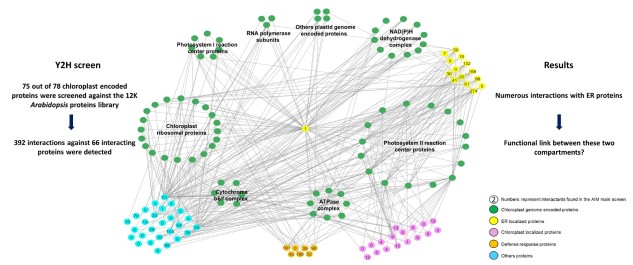
In eukaryotes, the endosymbiotic origin of cells and the inherent separation of intracellular compartments necessitate efficient communication between organelles and the nucleus. Evidences suggest that the endoplasmic reticulum (ER) and chloroplasts are biochemically connected, enabling, among other processes, the exchange of hydrogen peroxide (H2O2). This compound can also induce the formation of plastid stromules (stroma-filled tubular protrusions extending from the chloroplast main body) to carry signals to the nucleus. These signals lead to the activation of retrograde pathways and, if ROS production is high enough, to cell death.
25/09/2023
